(07/05/07) Roger Y. Tsien : Fluorescence and Imaging
by L.S. The Great Marduk.
As a graduate student at Cambridge in the late 1970s, Roger Y. Tsien measured intracellular calcium levels by injecting a calcium binding protein through the cell membrane – a very laborious process. Aside from being tedious, this method usually damaged the very cells being studied. Tsien eventually came to circumvent this problem by developing a dye that can make itself undetectable to the cell so that it can pass safely through the membrane without having to be injected. Since then, many changes have developed in the realm of intracellular imaging.
Cell visualization is an important tool in biology related fields. Understanding the mechanism of different processes inside the cell or identifying which gene codes for which protein requires a way to visualize the cell. By labeling certain parts of a cell with some kind of marker label, we can study in detail certain mechanisms or understand the internal structure of biological cells. However, traditional approaches involved methods detrimental to the cells being studied as Tsien demonstrated when he was a graduate student. Thus, a new method of marking cells would greatly improve cell visualization.
In the early 1990s, Tsien took the green flurescent protein GFP, derived from the jellyfish Aequorea Victoria, and reengineered it to emit colors ranging from blue to yellow. During this period, GFP became popular after a breakthrough by Douglas Prasher when he cloned the gene for the protein. Martin Chalfie then demonstrated that expression of the gene in organisms other than the jellyfish also creates fluorescence. Thus, the gene contains all the information necessary for the posttranslational synthesis of the chromophore, which is responsible for its color, without needing any jellyfish-specific enzymes. Attaching this protein to any other proteins allow scientists to visualize its location in the cell and thus, recombinant proteins such as GFP quickly outdated the traditional techniques.
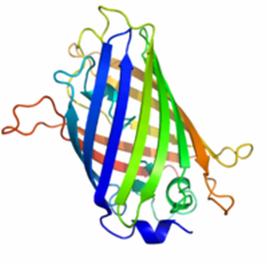
Figure 1 shows the general structure of GFP. It is composed of 238 amino acids and fluorescece green when exposed to blue light. It consists of an 11 strand beta-barrel with a single alpha helical strand containing the chromophore running through the center. The barrel permits chromophore formation while protecting it from the surrounding environment. GFP is usually useful as a reporter of gene expression but one major drawback is its 238 amino acids. This is troublesome because its large size might affect the protein being studied. Additionally, GFP will not fluorescece right away as it will first have to be express. Also, it can only be bind at the C or N-terminus.
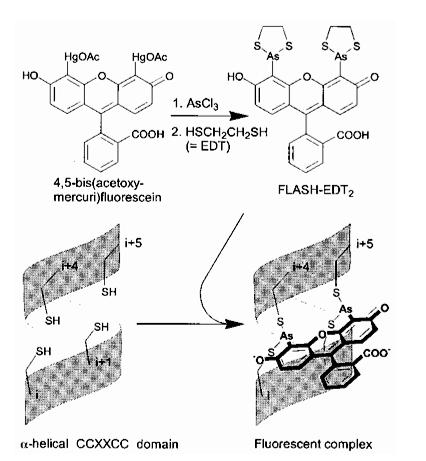
The Tsien lab at UCSD has mainly focused on fluorescence microscopy as a way to understand biological processes. Other small fluorescent proteins such as fluorescein isothiocyanate (FTIC) are usually toxic to the live cell. GFP and its derivatives, in comparison, are much less harmful when illuminated in live cells. However, as stated, one of its drawbacks is its size. Thus, Tsien had developed a smaller fluorescent protein known as FlAsH and ReAsH for use in cell visualization. Figure 2 depicts the synthesis of FlAsH. FlAsH is a membrane-permeant biarsenical dye with the advantage of being a much smaller tag in comparison to GFP. Thus, it can permeate cells without having to be injected and the smaller size makes it less perturbative to the protein being studied. It has relatively few binding sites in mammalian cells but will bind to the designed tetracysteine motif. Thus, it is much more specific than GFP. It also has the added bonus of being non-fluorescent until it binds to its target, whereupon it becomes strongly fluorescent. This is an improvement over GFP because it cuts out the delays time required for the maturation of GFP.
Dyes such as FlAsH and ReAsH usually binds to genetically encoded tetracysteine motifs expressed in living cells. However, it was soon noted that spontaneous nonspecific background staining could prevent detection of weakly expressed or dilute proteins. This problem could be solved if the affinity of the dye to the tetracysteine motif could be increased. This will allow for the fluorescent dyes to be more specific to the protein being studied instead of non-specifically bonding itself to the background of the cell. Thus, improvement to the affinity of the tetracysteine site is the focus of the first paper.
In the paper “Mammalian cell-based optimization of the biarsenical-binding tetracystein motif for improved fluorescence and affinity,” Tsien et al. attempted to improve ReAsH affinity by optimizing the tetracysteine sequence. In the earliest designs, partial reduction of the background fluorescence was achieved by increasing the concetration of the dithiols 1,2-ethane-dithiol (EDT) or 2,3-dimercaptopropanol (BAL) in washes to remove the thiol-dependent background or by including non-fluorescent dyes to block hydrophobic binding sites. This enhanced the affinity and contrast the proteins fused to FlAsH. In order to further improve affinity, tetracysteine sequence would have to be improved. This was done by developing a retrovirally transduced mammalian cell-based approach. Fluorescence-activated cell sorting (FACS) was used to select for the optimal residues surrounding the tetracysteine motif.
The first library, ReAsH Retroviral Library 1 (RRL1), was created by ligating a semi-randomized oligonucleotide cassette to the C terminus of the green fluorescent protein (GFP). This library was then introduced to NIH3T3 cells, which were stained with ReAsH. Thus, the oligonucleotides would assemble into a new sequence. The sequence would produce a tetracysteine motif where ReAsH will bind. Through fluorescence resonance energy transfer (FRET) between GFP attached to the nucleotides and ReAsH bound to the tetracysteine motif, Tsien was able to sort out which sequence improved the binding affinity of the tetracysteine site. Figure 3 showed the possible locations for the new nucleotides.
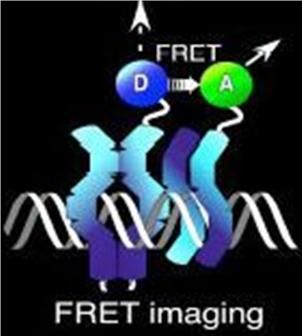
Normally, an excited fluorescent molecule such as GFP dispose of the energy from absorbed protons by emitting a photon of slightly longer wavelength (less energy). That is why GFP fluoresceces green when exposed to blue light. With FRET though, the excited protein molecule becomes a “donor” and passes the energy directly to a nearby molecule (acceptor) without emission of a photon, and thus, exciting the acceptor. The acceptor can now decay to its ground state by fluorescence. FRET is only possible when the donor and acceptor are close to each other (within 1 to 50 A depending on the donor/acceptor). Figure 4 depicts FRET.
The FRET efficiency is inversely proportional to the sixth power of the distance between the donor and acceptor. Thus, very small changes in the distance between donor and acceptor register as very large changes in FRET ratio. This is measured as the fluorescence of the acceptor molecule when the donor is excited. That is why in this experiment, Tsien can select for the sequence that increases tetracysteine affinity by measuring the fluorescence of ReAsH and sorting it through FACS. Thus, if specific ReAsH binding occurred, the fluorescence of ReAsH should increase while the fluorescence of GFP should decrease.
The first-generation tetracysteine sequence showed varying levels of FRET after washing with dithiols. This suggests that different amino acid combinations near the tetracysteine can modulate fluorescence properties of the complex. RRL1 cells exhibiting FRET were collected and expanded. Three additional rounds of sorting were done, each time increasing the selection pressure by increasing the dithiol concentration during washing. Sequence analysis of the isolated clones showed ten new tetracysteine sequences. The results confirmed the consensus sequence CCPGCC with PG in between the tetracysteine.
In order to further optimize the ReAsH binding tetracysteine peptide, a new library, RRL2, was devised whereby PG was fixed as the internal residues while the three external residues on either side of the tetracysteine were randomized (XXXCCPGCCXXX). The abbreviation XXX#XXX will now be used where #=CCPGCC.
The same method from round 1 of selection was used and three new sequences were converged upon, HRW#KTF, FLN#MEP, and YRE#MWR. Further selection showed that FLN#MEP and HRW#KTF provided the best sequence for improved tetracysteine affinity. The conclusion of the paper stated that Tsien et al did not have a complete molecular explanation of the higher affinity of the final improved tetracysteines. However, alanine scanning suggests that large aromatic residues are useful, possibly shielding the biarsenical-tetracysteine complex from competing dithiols. Also, charge did not seem to provide a explanation because HRW#KTF has a charge of +2 to +3 while FLN#MEP has a -1 charge while the residues contributing to the high dithiol resistance were neutral. They believe that further understanding of the affinity and properties of the complexes will likely require high-resolution structures. However, they prefer FLN#MEP over HRW#KTF because the former gives higher quantum yields.
The second paper “Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy” also uses FRET between ReAsH and GFP to perform the experiment. In the paper, Tsien used FRET to study the reconstitution of the Golgi apparatus during cell division. The Golgi apparatus is an organelle found in typical eukaryotic cells whose primary purpose is to process and package macromolecules synthesized by the cell. In the interphase stage, the Golgi apparatus has a ribbon-like shape joined together by a tubular network and anchored in the centrosomal region of the cytoplasm. During cell division, it undergoes fragmentation and their components disintegrate and diffuse throughout the cell in the form of small clusters sized below the resolution of the light microscope. These clusters are generally referred to as the “Golgi haze.” Since these clusters are so small, their true nature are not really known but it is believed that they coalesces around the endoplasmic reticulum (ER). Afterwards, the Golgi apparatus reconstitute through a series of poorly understood events. Fusing Golgi-resident enzymes with fluorescent proteins could perhaps shed light on its behavior during cell division.
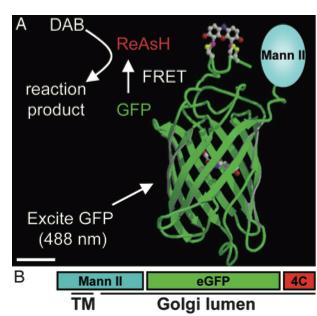
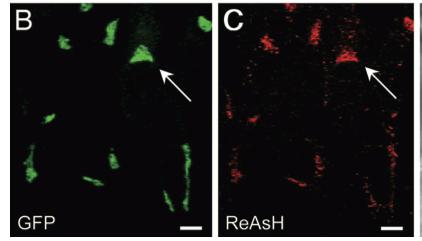
As stated before, the 238 amino acids of GFP can disturb the behavior of the molecule being studied if the molecule in question is relatively small. This is the case with the Golgi haze. Thus, an alternative method was needed. This experiment used the singlet oxygen generated by the photoexitation of ReAsH to show the location of the Golgi haze during cell division. In the experiment, CFP (cyan color) was fused to alpha-mannosidase II (MannII), a Golgi resident enzyme. CFP was used rather than GFP because of the spectral overlap of FlAsH and GFP (both fluorescing green). CFP was attached to the carboxyl terminus of a tetracysteine containing peptide. Thus, MannII will always be around particles of the Golgi apparatus while it is attached to GFP with a binding site for ReAsH. A image of this can be seen in Figure 5. In the experiment, MannII-CFP-4C was expressed in HeLa cells, where it would localize in the Golgi apparatus. The cell has also been stained with ReAsH. However, the tetracysteine residues in MannII-CFP-4C are exposed to the oxidizing environment of the Golgi lumen and would not be avaible for ReAsH because binding of the biarsenicals to the tetracysteine tag requires the cysteine residue to be completely reduced. Only when the membrane-permeant reducing agents tributylphospine (TBP) or triethylphosphine is (TEP) administered will ReAsH be allowed to label MannII-CFP-4C. Thus, that is how they control the timing of ReAsH photoexitation.
When TBP or TEP is administered, ReAsH will be allowed to bond to the tetracysteine motif attached to CFP. This is attached to MannII, which will be localized around the Golgi haze. Figure 6 shows the cell before and after ReAsH bonding. FRET between CFP and ReAsH will cause a photoexitation of ReAsH, which will generate a singlet oxygen. The singlet oxygen will locally polymerize diaminobenzidine (DAB) stained throughout the cell into a fine precipitate by photoconverting it. The golgi localized precipitate will be rendered electron-dense through treatment with osmium tetroxide and will then go through high-resolution analysis by electron microscope.
Through this imaging technique, Tsien et al. found that the Golgi clusters did not localize to the ER or other organelles at metaphase and anaphase as previously believed. The fragments appeared to retain autonomy throughout mitosis. The imaging also found that four clusters of vesicles and tubular structures were formed at the edge of the midbody and distal to the midbody with one pair in each daughter cell. The twin clusters in each daughter cell then gradually fuse back together and form a single Golgi apparatus in a pericentrosomal location.
While the experiment was a success in showing the reconstitution of the Golgi apparatus, it achieved a bigger purpose of expanding the role of fluorescent proteins beyond the use of just fluorescence microscopy. Tsien et al. demonstrated a more versatile tagging system and extended its applicability to intracellular sites inside of oxidizing environments. Such capabilities can allow scientists to study other intracellular organelle systems.
The third paper “Control of mammalian translation by mRNA structure near caps” studied the effects of hairpin location, thermal stability, and GC content on the translation efficiency. Hairpins are small RNA aptamer motifs inserted into the 5’ UTR of mRNAs to repress protein production upon ligand addition. RNA structures in the 5’ untranslated region (UTR) are important to the regulation of translation by affecting ribosomal recruitment and positioning at a favorable initiation codon. Previous studies by M. Kozak demonstrated that hairpins with thermal stability of -30 kcal/mol had no effect on translation while hairpins of -50 kcal/mol reduced translation by 85%-95%. Kozak also showed that stem-loop-cap-to-hairpin distance plays a role in translation efficiency. In the previous studies, structures of -30 kcal/mol hairpins situated 12 bases from the cap reduce translation. However, when the distance was increased to 52 base pairs, there was no effect.
The experiment performed by Tsien et al expands upon the previous work to investigate in better detail how RNA structures affect translation efficiency in cells. As usual with all of Tsien’s experiments, fluorescent proteins were used. In this experiment, a translation reporter was constructed by allowing manipulation of 5’ UTR sequences upstream of a GFP protein. The original enzyme sites encompassing a transcriptional start site were cloned. The site where RNA transcripts was initiated was reverified with additional hairpin constructs using RNase protection. Adding an RFP protein on the same vector normalized for experimental variability. Transfection of this vector into Cos7 cells resulted in green and red emissions indicating translation. Measuring the ratio of GFP to RFP with fluorescence microscopy is a measurement of translation efficiency.
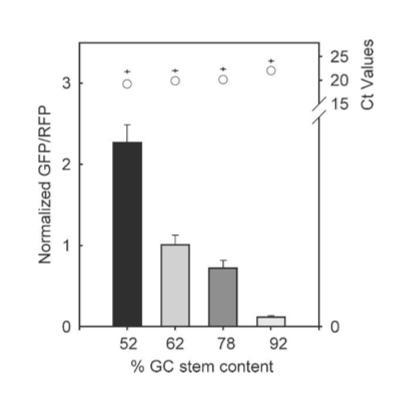
The results from figure 7 showed that translation efficiency is increased as the hairpin position increased. However after a distance of 12 base pairs, the translation efficiency starts to decrease indicating that the optimal distance for hairpin position is 12 base pairs. For thermal stability, the average thermal stability values were taken for eight different distances for the hairpin position. Translation efficiency is optimal when the thermal stability is between 10 to approximately 22 kcal/mol, after which it quickly decreases. Tsien et al also discovered that translation efficiency was dependent upon RNA-hair GC stem content. The hairpins’ content was designed with different amounts of GC within the structure while maintaining a position at +4 and thermal stability of -30 kcal/mol. It was shown that between 52% and 92% GC content, there was an 18-fold decrease in translation efficiency. Thus, they concluded that natural hairpin structures should also regulate translation efficiencies depending on the three different factors studied in their experiment.
The fourth paper entitled “Evolving Proteins in Mammalian Cells Using Somatic Hypermutation” was a change of pace from the previous papers. For this paper, a new method was described to mutate target genes through somatic hypermuation (SHM) by evolving proteins directly in living mammalian cells. Unlike the previous papers, this paper had more of a recipe-like feel because it did not seek to specifically answer a question. However, it retained some connection to the other papers by involving a monomeric red fluorescent protein (mRFP1.2).
To traditionally evolve new protein properties, gene libraries normally had to be expressed using in vitro methods. These methods include mutagenesis and in vitro gene recombination. The genes are then introduced into cells where selection or screening can be done. The next round requires another in vitro mutagenesis, which can be extremely tedious when done iteratively. Also, the size of the gene library that can be introduced into the cells is limited by the efficiency of transformation, transfection, or infection. If there was a way to create genetic diversity directly in living cells, then these problems could be circumvented and allow for larger protein sequence spaces to be sampled for desired protein properties.
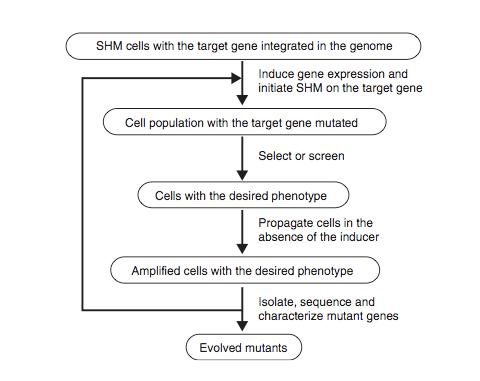
Figure 8 shows the schematic protocol for evolving proteins directly in mammalian cells through SHM. SHM is a process through which the B lymphocytes in the immune system mutate the Ig genes to produce antibodies when lymphocytes are activated by antigens. SHM then introduces point mutations into the rearranged V regions of the Ig genes at a rate of approximately 1 x 10-3 mutations per base pair per generation. In the paper, Tsien et al showed that SHM could introduce multiple mutations throughout a target gene and that beneficial mutations could be retained after rounds of evolution to generate useful phenotypes and properties. They chose to evolve mRFP1.2 in Ramos cells so that the mutant, dubbed mPlum, will have a far-red emission.
The host Ramos cell is a B-cell line that hypermutates its Ig V gene constitutively during culture. The target gene is integrated into the genome by expressing it under the control of the inducible Tet-on promoter and addition of the inducer doxycycline. The mutated genes are then expressed and are selected for the desired properties. The cells are then subjected to another round of mutation and selection. Since this is all done in vivo, the process can be iterated much more easily for numerous additional rounds until the cell exhibited the desired properties.
For the evolution of mRFP1.2 to mPlum, FACS was used to seek mRFP mutants with far-red emission. It took 23 rounds of SHS and FACS to evolve mPlum. The final product had an emission maximum at 649 nm and an increased photostability compared to the parental mRFP. Sequencing mPlum showed 7 substitutions out of 225. The group found that SHM could introduce multiple mutations into the target gene each round and that it could sample a larger sequence space of proteins, thus, showing a nice improvement over the previous in vitro method.
The final paper looked at was “Systems Analysis of PKA-Mediated Phosphorylation Gradients in Live Cardiac Myocytes.” It focused on the dynamics between cyclic AMP (cAMP) and its cAMP-dependent protein kinase (PKA). In the heart, compartmentation appears to contribute to functional differences between beta1- and beta2- adrenergic signaling and other stimuli that increase cAMP and thus, is important for understanding the role of beta-adrenergic signaling in the development and treatment of heart failure. Short term beta-adrenergic signaling increases heart contractility while prolonged exposure to beta1-adrenergic induces apoptosis.
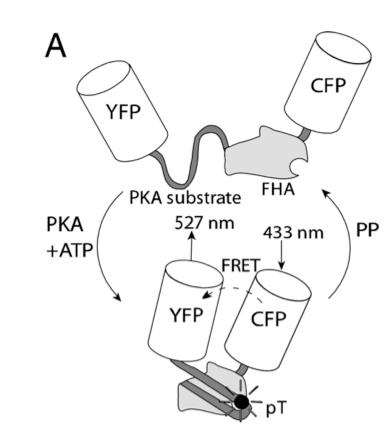
In this experiment, the researchers used fluorescent reporters to understand the signaling mechanism contributing to spatially heterogeneous PKA-mediated phosphorylation. A-kinase activity reporter 2 (AKAR2) has been shown to be a good indicator of PKA-mediated phosphorylation. To study compartmentation in heart cells, AKAR2 was transfected into neonatal rat cardiac myocytes. Each end of AKAR2 had been attached to fluorescent reporters. In this case, they are yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP). If phosphorylation occurred, AKAR2 would fold in such a way that FRET will occur between YFP and CFP. This is shown in figure 9. By studying the emission ratio yellow/cyan fluorescence of AKAR2, the researchers can follow the phosphorylation in the cardiac myocytes.
To activate phosphorylation, all that is needed is UV light. Distributed throughout the cell are 4,5-dimethoxy-2-nitrobenzyl (DMNB-cAMP) that are caged along with PKA. To uncage DMND-cAMP, UV light is exposed to the cell, which then activates the PKA. This causes phosphorylation, which then changes the conformation of AKAR2 and can be measured through cell imaging as previously described.
Tsien et al found that cAMP accumulation was rate-limiting in PKA-mediated phosphorylation in response to beta-adrenergic in this system. AKAR2 dephosphorylation rates revealed that saturation of PKA with cAMP could delay substrate dephosphorylation, indicating a capacity for cAMP synthesis greatly exceeding the requirements for PKA activation. They also found that PKA-mediated phosphorylation in response to local uncaging of DMNB-cAMP confirms that cAMP compartmentation can facilitate gradients in phosphorylation, a necessary step toward mediating functional diversity of PKA signaling.
The five papers discussed provide a good overview of the kind of work that Roger Tsien does at UCSD. After going through the papers, there are a couple of experiments that can be a follow through to some of his work. At the end of the paper “Mammalian cell-based optimization of the biarsenical-binding tetracystein motif for improved fluorescence and affinity,” it was concluded that the researchers did not have a complete molecular explanation of the higher affinity of the improved tetracysteines. The alanine scanning suggests that it might have something to do with the large aromatic residues. They might have possibly shielded the biarsenical-tetracysteine complex from competing dithiols.
A good follow up experiment would be to synthesize a tetracysteine motif that has large aromatic residues and another that does not. Using FRET again, the researchers can then see if there is a stronger affinity for the tetracysteine with the large aromatic residues. This can confirm that it is the large aromatic residues that are the reason why HRW#KTF and FLN#MEP were good tetracysteine sequences.
With the methods of tagging introduced in the paper “Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy,” its applicability can be extended to other intracellular sites that has an oxidizing environment or that is too small to be resolved by the light microscope. Aside from just imaging pathways and mechanisms, perhaps the technique can be used to develop drugs that use nano delivery. This way, researchers will be able to understand the specific pathway of such drug delivery.
Figure 1. A ribbon model of the green fluorescent protein (GFP)
Figure 2. Synthesis of ReAsH
Figure 3. Possible location of the new nucleotides during round 1 synthesis of a new tetracysteine sequence
Figure 4. A diagram of fluorescence resonance energy transfer (FRET)
Figure 5. A combinatorial tag used for identifying the location of the Golgi haze
Figure 6. Imaging of the Golgi haze before and after photoconversion
Figure 7. Results of hairpin experiment
Figure 8. Schematic diagram of the protocol for evolving proteins directly in mammalian cells
Figure 9. Schematic of AKAR2, with FRET from CFP to YFP upon phosphorylation by PKA
References
Original Five Articles
1) Saucerman, Jeffrey J.; Zhang, Jin; Martin Jody C.; Peng, Lili X.; Stenbit, Antine E.; Tsien, R.Y.; McCulloch, Andrew D., Proc. Natl. Acad. Sci. 2006, 103, 12923-12928
2) Wang, L.; Tsien, R.Y., Nature Protocols 2006, 1, 1346-1350.
3) Guido M. Gaietta; Ben N. G. Giepmans; Thomas J. Deerinck; W. Bryan Smith; Lucy Ngan; Juan Llopis; Stephen R. Adams; Roger Y. Tsien; Mark H. Ellisman, Proc. Natl. Acad. Sci. 2006, 103, 17777–17782
4) Jeremy R. Babendure; Jennie L. Babendure; Jian-Hua Ding; Roger Y. Tsien, RNA 2006, 12, 851–861
5) Brent R. Martin; Ben N.G. Giepmans; Stephen R. Adams; Roger Y. Tsien, Nature Biotechnol 2005, 10,1308-1314.
Additional Resources
6) Hauser, C.T.; Tsien, R.Y., Proc. Natl. Acad. Sci. 2007, 104, 3693–3697
7) Griffin, B. Albert; Adams, Stephen R.; Tsien, R.Y., Science. 1998, 281, 269-272
8) Tsien, R.Y., Annu. Rev. Biochem. 1998, 67, 509–44 Delete the arrows from this line


Enable comment auto-refresher